Novel molecular subtypes and EBV-associated Reprogrammed Regulatory Epigenome in Nasopharyngeal Carcinoma - 03 November 2022
Chow LKY*, Chung DL*, Tao L, Tung SY, Ngan RKC, Ng WT, Lee AWM, Yau CC, Kwong DLW, Lee VHF, Lam K, Liu J, Chen H, Li ML#, Dai W#. Epigenomic Landscape Study Reveals Molecular Subtypes and EBV-associated Regulatory Epigenome Reprogramming in Nasopharyngeal Carcinoma. EBioMedicine 86, 104357 (2022).
(* Joint first authors # Co-corresponding authors)
Researchers from the Department of Clinical Oncology, School of Clinical Medicine, LKS Faculty of Medicine, the University of Hong Kong discovered a novel epigenomic subtype of Epstein-Barr (EBV) nasopharyngeal carcinoma (NPC), and EBV-associated reprogrammed regulatory epigenome relevant to immunosuppressive EBV-specific communications in the tumor microenvironment (TME). These findings provide novel insights into NPC pathogenesis and highlight the interplay of host factors and viral-associated epigenetic reprogramming. The research has been accepted in EBioMedicine.
Background
Nasopharyngeal carcinoma is highly endemic in Southeast Asia, including Hong Kong and Guangdong Province in Mainland China. Strikingly, EBV can be detected in more than 90% of patients in Hong Kong. Pioneer epidemiology and genomic sequencing studies have provided substantial evidence that host genetics, EBV and environmental factors are critical risk factors in NPC. Previous epigenetics studies showed that predominant genome-wide DNA hypermethylation was a unique feature that distinguishes EBV-positive NPC (EBV+ NPC) from other cancer types. EBV can dysregulate epigenetics machinery. Although the importance of epigenetics has been highlighted in NPC, chromatin accessibility, critical for regulatory epigenome, has not been comprehensively profiled. Whether and how EBV induces epigenetic changes, or whether host factors are involved in EBV-associated epigenetic changes remain largely elusive.
Research Findings
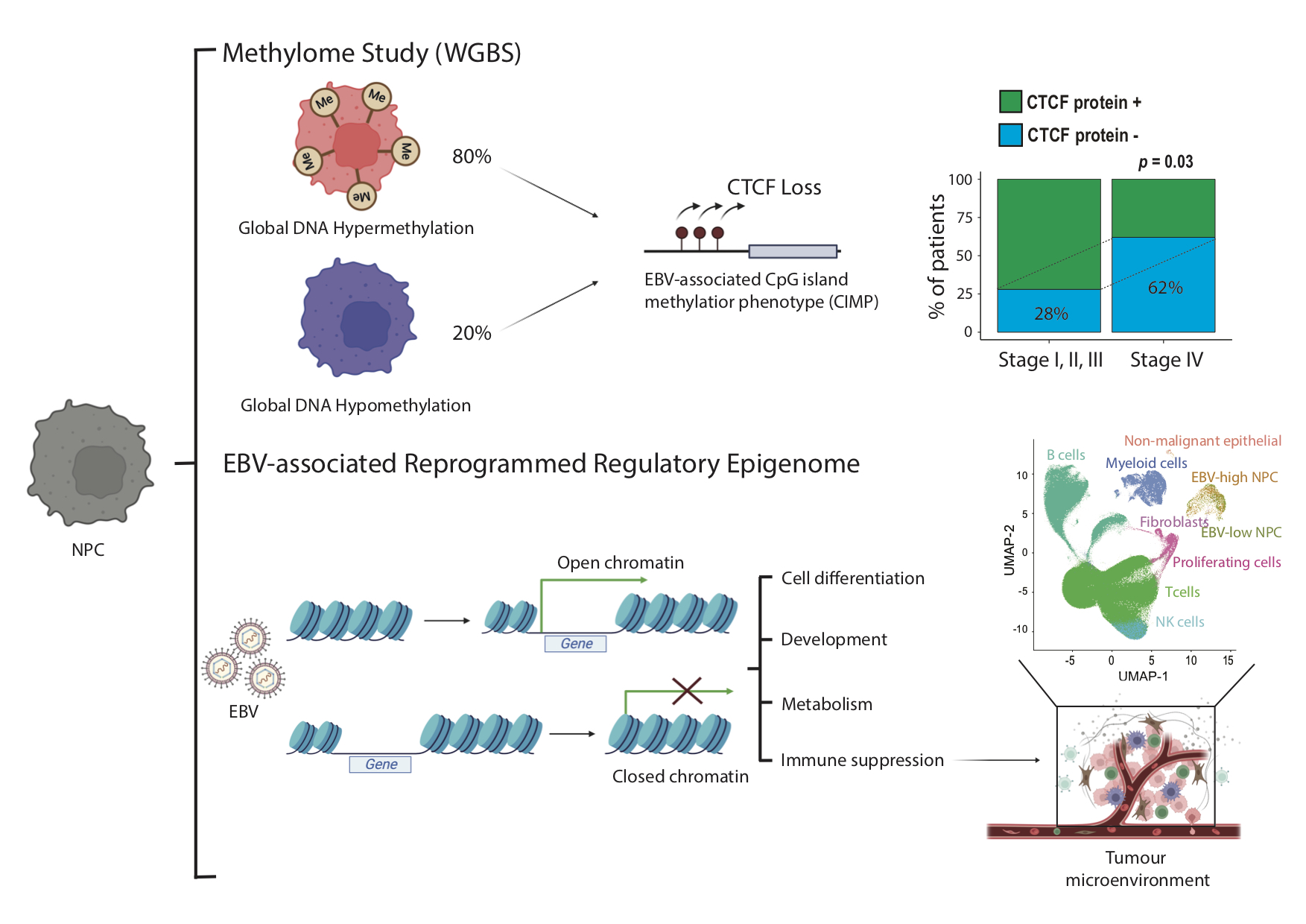
This epigenomic landscape study has discovered an epigenome subtype of EBV+ NPC characterized by global DNA hypomethylation in a subset of clinical specimens and the NPC cell line model, in addition to the known globally DNA hypermethylated EBV+ NPC. The team provides evidence that EBV cannot solely shape the global methylation pattern in NPC and the host factor CTCF may play an important role for epigenetic dysregulation in NPC. The reprogrammed regulatory network with the switching of transcription factor binding and altered chromatin accessibility was revealed in EBV+ NPC, contributing to the upregulation of CD74, which mediates EBV-specific cell-cell communications in the tumor microenvironment of NPC patients.
Significance of the study
It has been proposed that global hypermethylation is a critical step in pathogenesis in EBV+ NPC, discovery of the globally hypomethylated molecular subtypes reveals the new mechanisms underlying EBV+ NPC tumorigenesis. When global hypomethylation occurs during NPC pathogenesis, whether it occurs as an alternative pathway in a subset of patients, and its potential prognosis value are critical for a comprehensive understanding of NPC. In addition to EBV, host factors are involved in epigenome reprogramming in NPC pathogenesis. The immunosuppressive cell-cell communications in the EBV+ NPC could be mediated by epigenome reprogramming. These findings highlight the importance of molecular subtyping in EBV+ NPC and EBV-specific communications as potential therapeutic targets and biomarkers in NPC.
About the research team
This research was co-supervised by Dr. Wei Dai, Assistant Professor and Prof. Maria Li Lung, Emeritus Professor of the Department of Clinical Oncology, School of Clinical Medicine, HKUMed. Dr. Ka Yue Larry Chow and Mr. Lai Shun Dittman Chung from the Department of Clinical Oncology, School of Clinical Medicine, HKUMed, are the co-first authors. Dr. Lihua Tao, Scientific Officer, Department of Clinical Oncology, School of Clinical Medicine, HKMed, provided assistance to the research.
The collaborators included Dr. Kui Fat Chan and Dr. Stewart Yuk Tung from Department of Clinical Oncology and Department of Clinical Pathology from the Tuen Mun Hospital, Hong Kong; Prof. Kai Cheong Roger Ngan, Prof. Tong Wai Ng Wai, Prof. Anne Wing Mui Lee, Prof. Dora Lai-Wan Kwong, Dr. Victor Ho-Fun Lee and Dr. Ka-On Lam from the Department of the Clinical Oncology, School of Clinical Medicine, HKUMed; Dr. Chun Chung Yau from Department of Oncology from Princess Margaret Hospital, Hong Kong; Prof. Honglin Chen and Dr. Jiayan Liu from Department of Microbiology, School of Biomedical Sciences, HKUMed.
About Department of Clinical Oncology
The Department of Clinical Oncology is an academic department specialized on full spectrum of non-surgical cancer services including radiotherapy, chemotherapy, targeted therapy and palliative care. The Department has two research laboratories including Laboratory of Cancer Molecular Genomics and Laboratory of Cancer Genetics and provides state-of-the-art cancer management at two hospitals (Queen Mary Hospital in Hong Kong and The University of Hong Kong-Shenzhen Hospital in China) in close collaboration with multiple disciplines.
Acknowledgment
This study was supported by the Hong Kong Research Grants Council grant (AoE/M-06/08), General Research Fund (17103218 and 17102619), seed funding for basic research (201611159158), and General Research Fund (17119618).
Molecular Subtypes based on the Mutational Signature for Nasopharyngeal Carcinoma
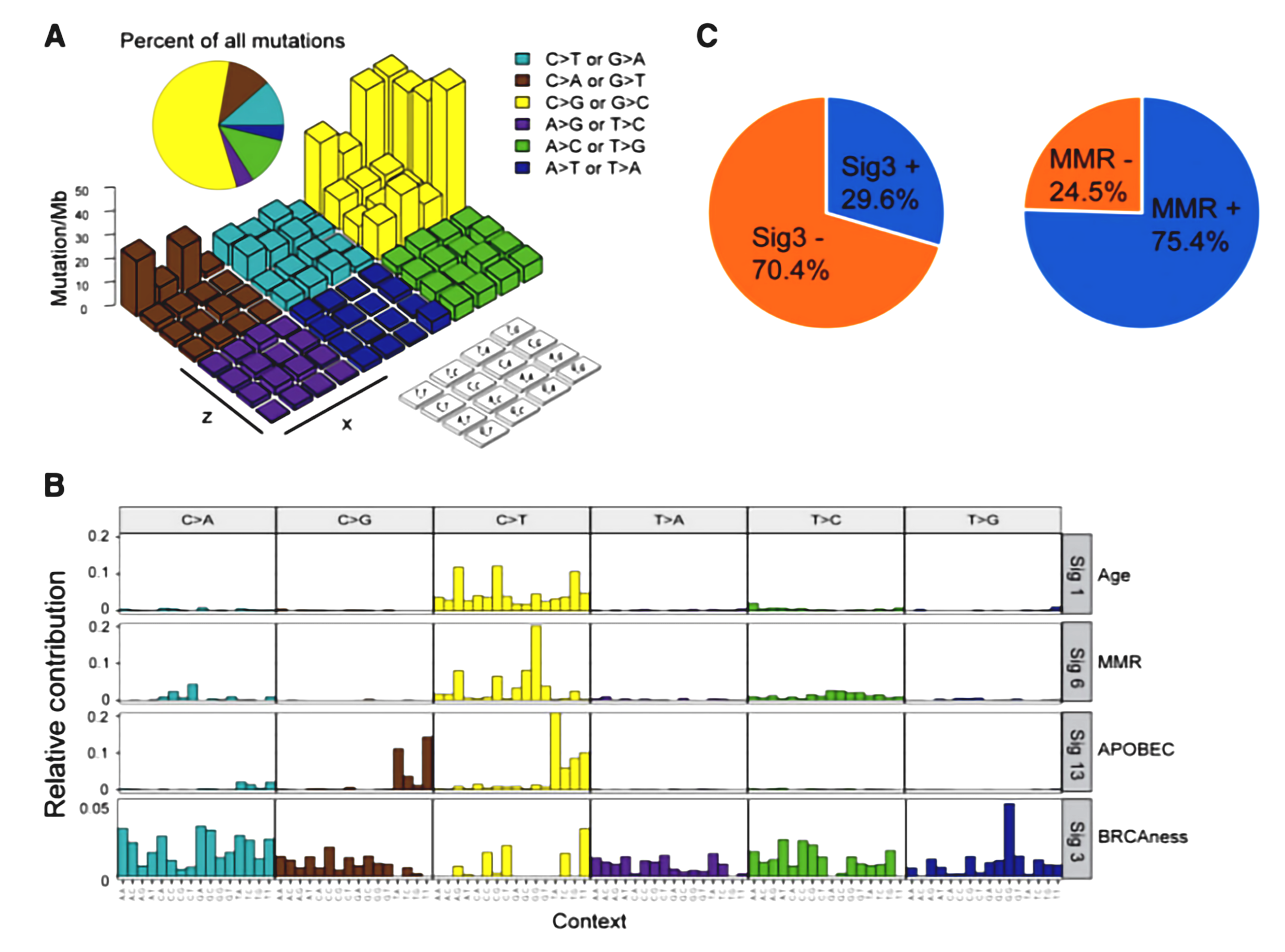
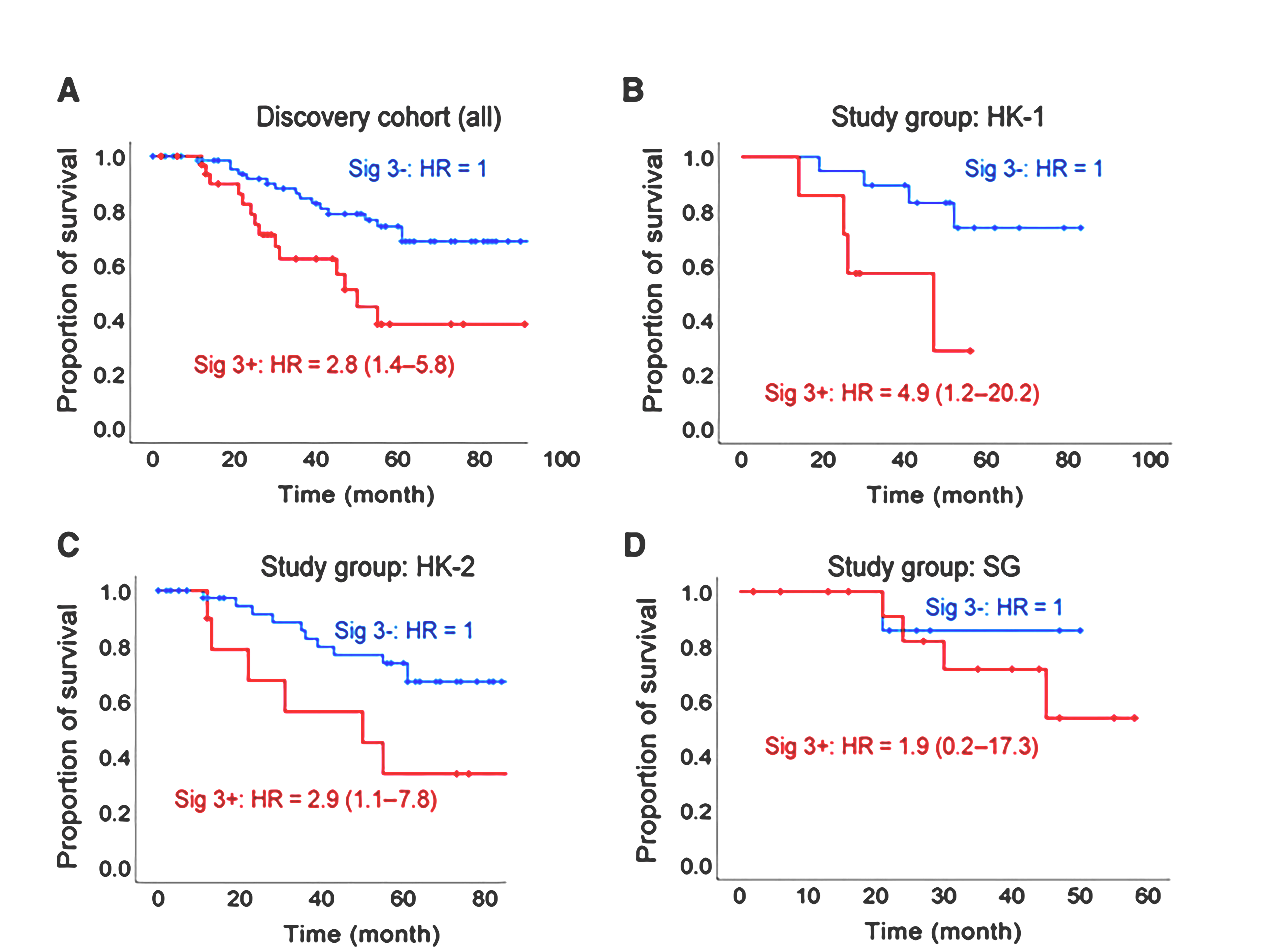
Dai W#, Chung DLS, Chow LKY, Yu VZ, Lei LC, Leong MML, Chan CKC, Ko JMY, and Lung ML#. Clinical Outcome-Related Mutational Signatures Identified by Integrative Genomic Analysis in Nasopharyngeal Carcinoma. Clin Cancer Res. 2020; 26(24).(# co-corresponding author)
In Nasopharyngeal Carcinoma (NPC), general consensus for treatment is to use radiotherapy (RT) alone for stage I disease, RT with or without concurrent chemotherapy for stage II and chemoradiotherapy (CRT) for advanced stage disease. However, 15-58% of the cases do not respond well to conventional treatment, and, thus, have poor clinical outcomes. There is a need for biomarkers to assist our understanding of the molecular basis of disease pathogenesis and progression and to aid clinical management in NPC. We have systematically examined the mutation signatures and evaluated their prognostic values in association with clinical outcome. The mutational signature relevant to homologous recombination deficiency (BRCAness) was discovered, which was unappreciated in NPC before. Importantly, independent prognostic values of the BRCAness signature and mismatch repair signature are now revealed. These data show the clinical importance of DNA repair pathways in NPC and their potential as prognostic and predictive biomarkers for future clinical studies.